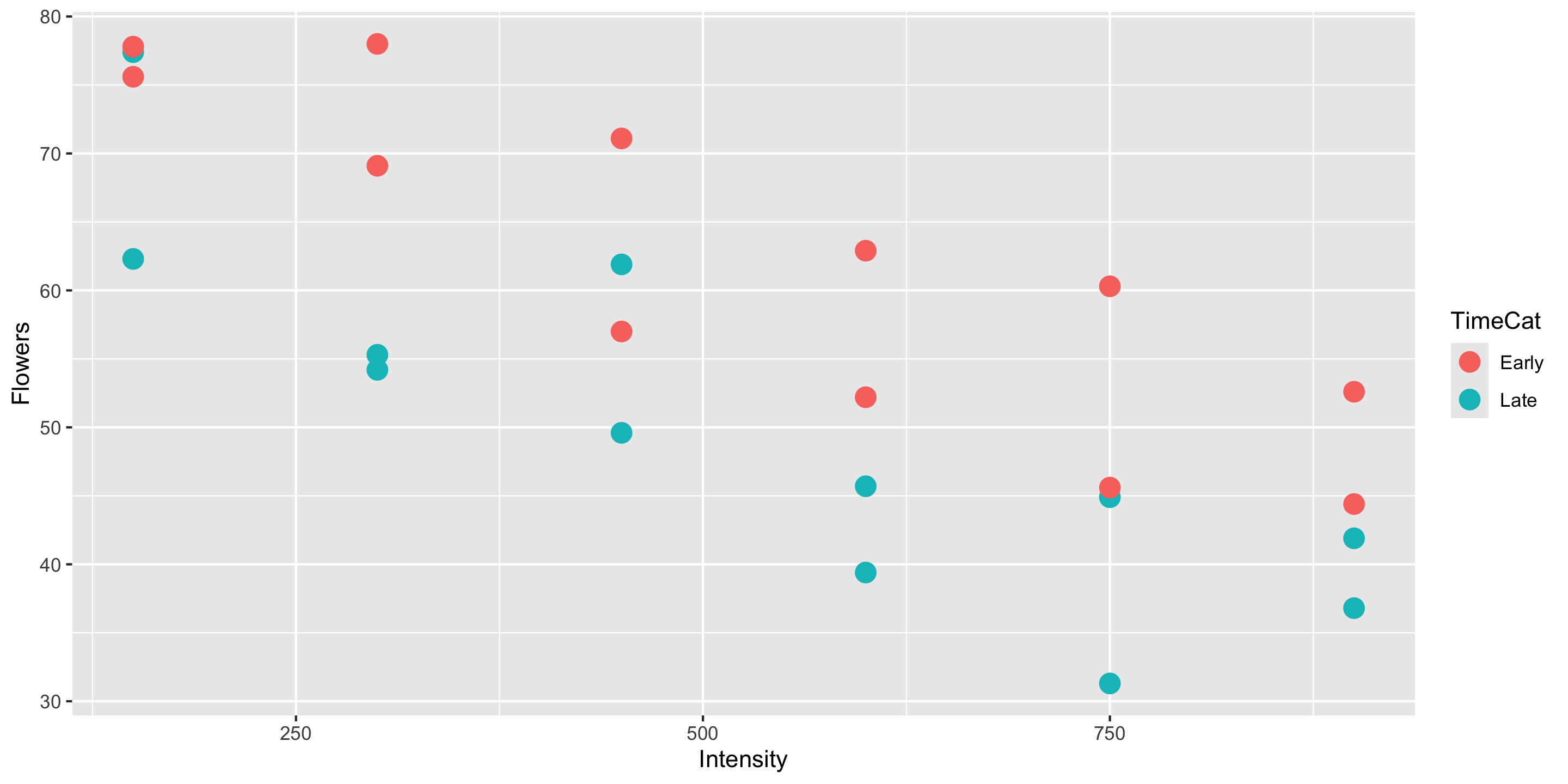
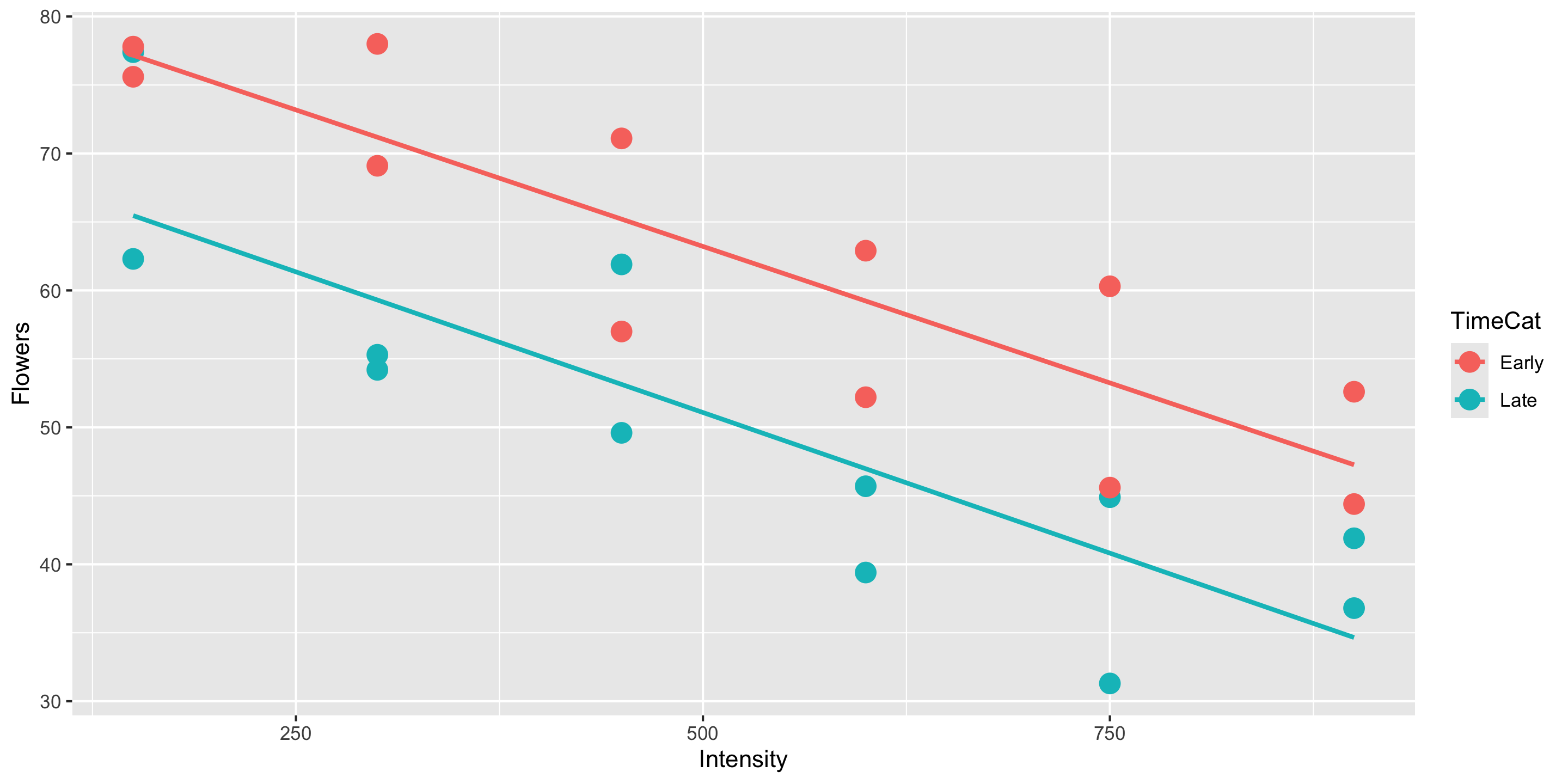
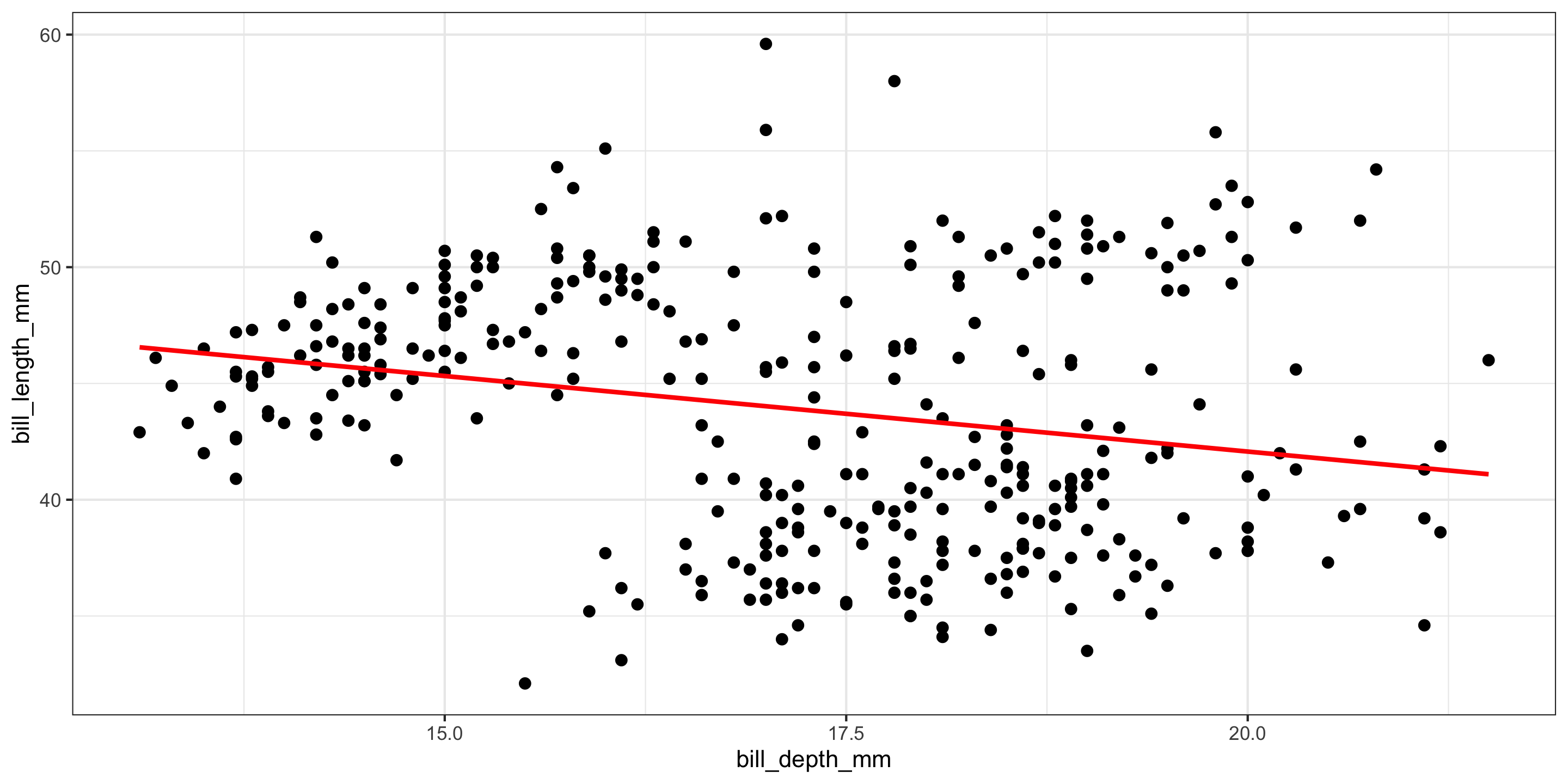
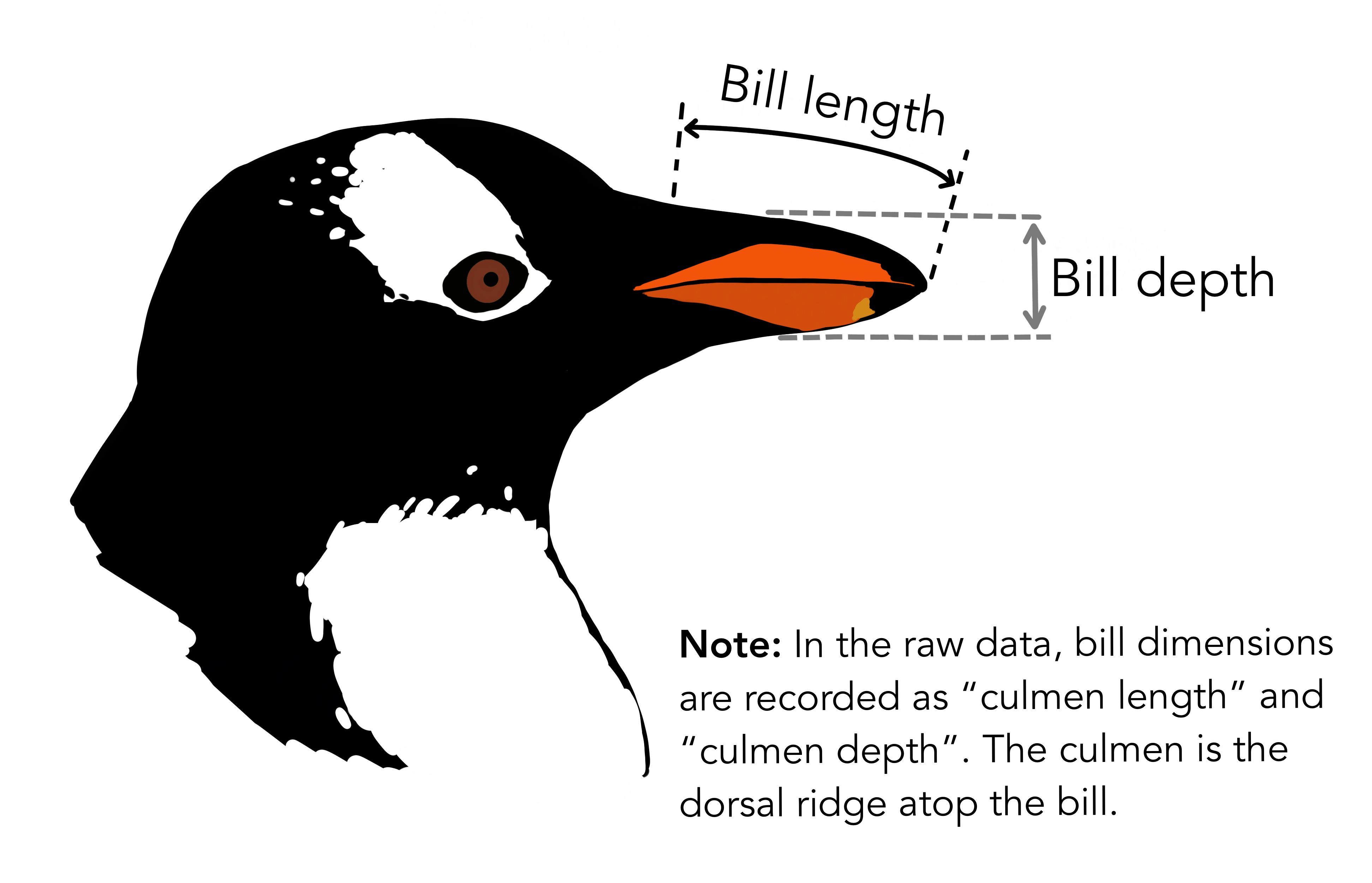
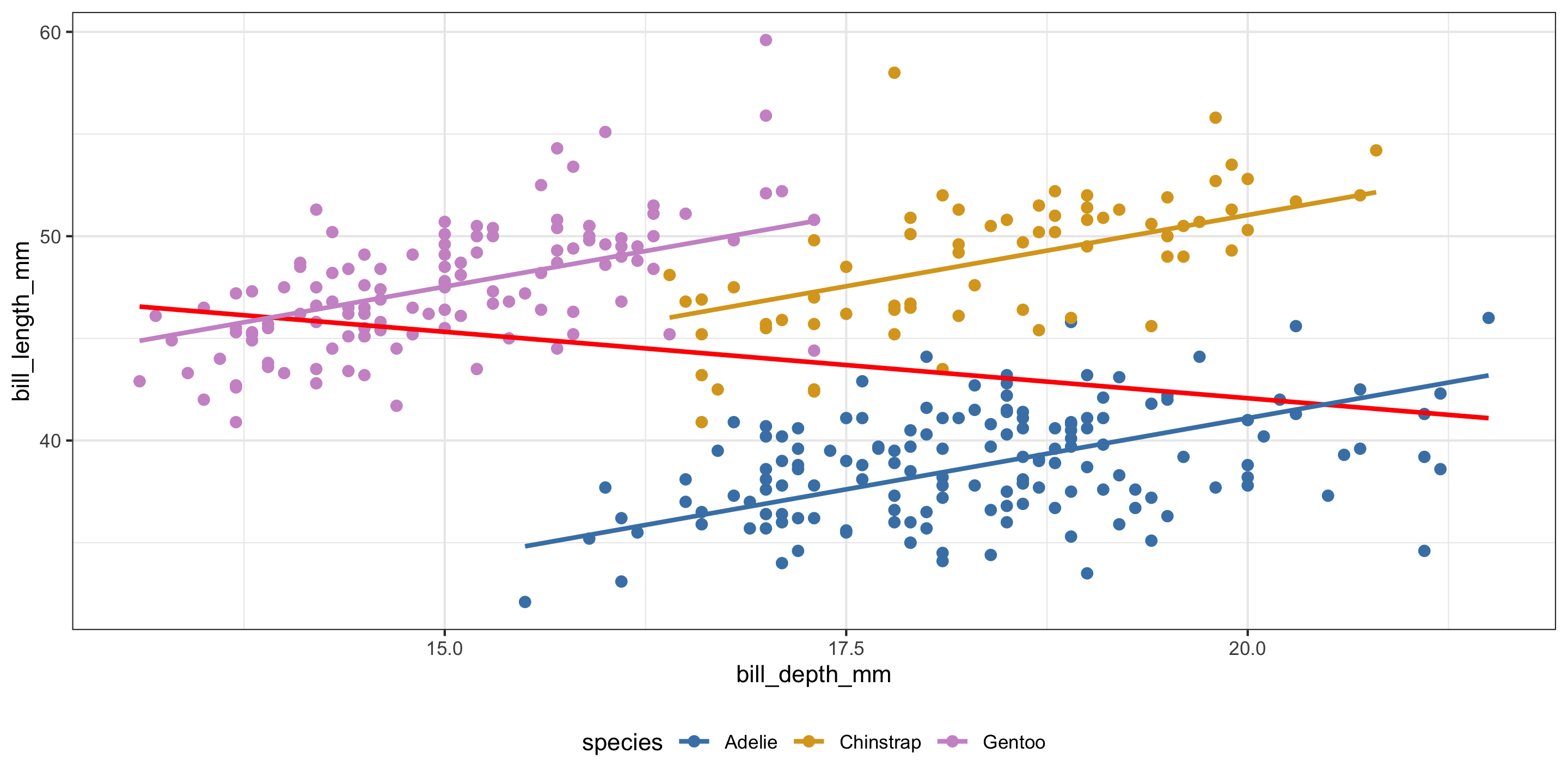
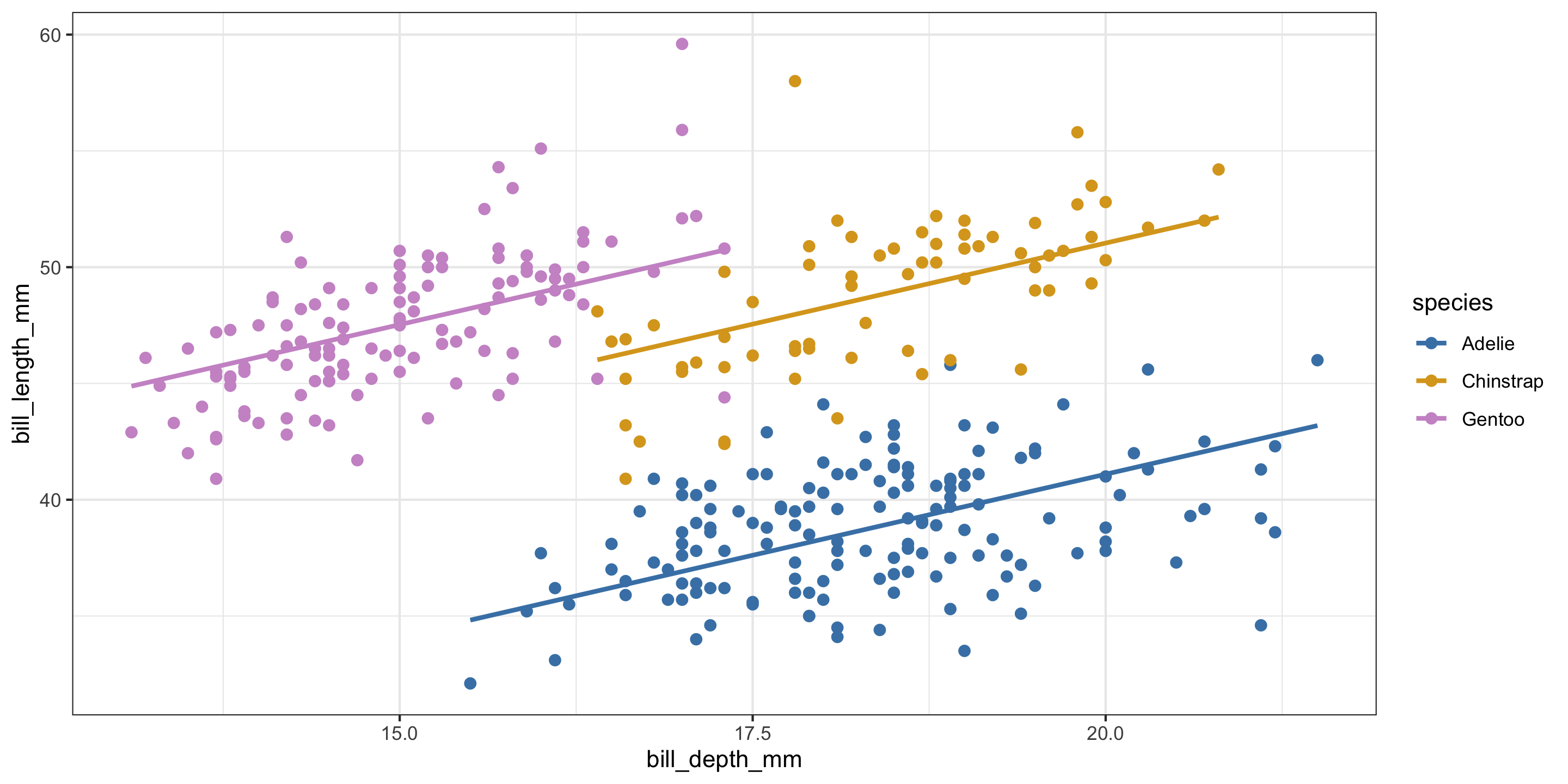
library(tidyverse)
library(Sleuth3)
data(case0901)
# Recode the timing variable
count(case0901, Time) Time n
1 1 12
2 2 12case0901 <- case0901 %>%
mutate(TimeCat = case_when(
Time == 1 ~ "Late",
Time == 2 ~ "Early"
))
count(case0901, TimeCat) TimeCat n
1 Early 12
2 Late 12